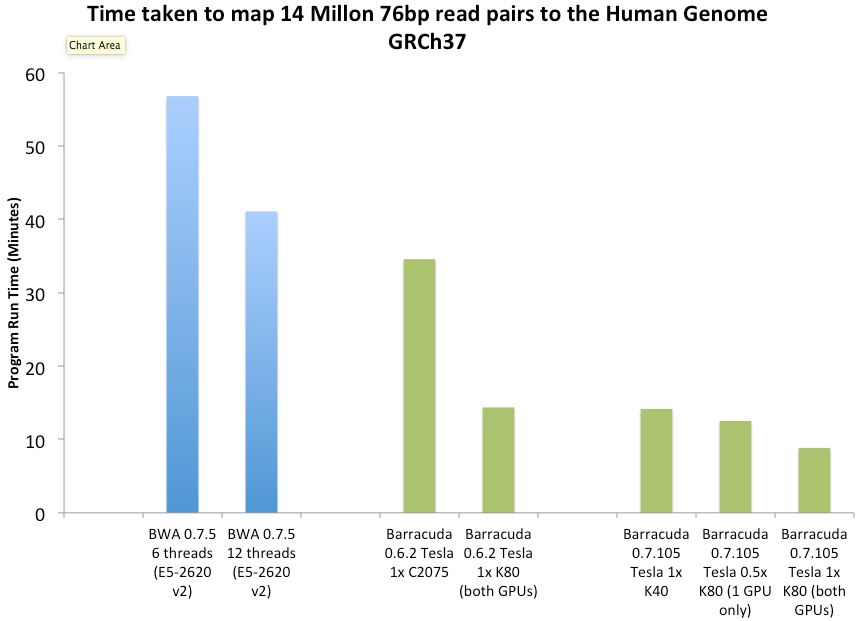
 The alignment throughput of 0.7.x vs BWA 0.7.5
The alignment throughput of 0.7.x vs BWA 0.7.5

Welcome to the BarraCUDA Project page
|
Started in 2009, the aim of the BarraCUDA project is to develop a sequence mapping software that utilizes the massive parallelism of graphics processing units (GPUs) to accelerate the inexact alignment of short sequence reads to a particular location on a reference genome. BarraCUDA can align a paired-end library containing 14 million pairs of 76bp reads to the Human genome in about 9 minutes from fastq files to SAM alignment using a NVIDIA Tesla K80. The alignment throughput can be boosted further by using multiple GPUs (up to 8) at the same time. Being based on BWA version 0.6 (http://bio-bwa.sf.net) from the Sanger Institute, BarraCUDA delivers a high level of alignment fidelity and is comparable to other mainstream alignment programs. It can perform gapped alignment with gap extensions, in order to minimise the number of false variant calls in re-sequencing studies. |
Software Features
|
|
|
Citations Please cite the following two papers when you use BarraCUDA: Langdon WB, Lam BY, Petke J, Harman M. (2015) Improving CUDA DNA Analysis Software with Genetic Programming. Proceedings of the 2015 Annual Conference on Genetic and Evolutionary Computation - GECCO '15 [DOI: 10.1145/2739480.2754652] Klus P, Lam S, Lyberg D, Cheung MS, Pullan G, McFarlane I, Yeo GSH, Lam BY. (2012) BarraCUDA - a fast short read sequence aligner using graphics processing units. BMC Research Notes, 5:27. [PMID: 22244497] Li H and Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics, 25:1754-60. [PMID: 19451168] |
* Measured using a system equipped with NVIDIA Tesla K80 and 2x Intel Xeon V5-2630v2 Processor.
The timings were based on the time taken for aligning the two libraries (-aln command) and also outputting in SAM format (-sampe command)
Last updated: 28 May 2015
|
Using genetic programming, v0.7.x now offers a >100% boost in speed compared to v0.6.2 Supports new BWTs built for BWA Version 0.6.x+ |
 The alignment throughput of 0.7.x vs BWA 0.7.5
The alignment throughput of 0.7.x vs BWA 0.7.5
 The alignment throughputs of 1 to 8x Tesla C2050s compared to equal number of x86 cores. (Klus et al., 2012)
The alignment throughputs of 1 to 8x Tesla C2050s compared to equal number of x86 cores. (Klus et al., 2012)